If you’re rare, do I care? A modeling tutorial
Preface
Description
This is a modeling tutorial for the supplement for Miller-ter Kuile et al. “If you’re rare, should I care? How imperfect detection changes relationships between biodiversity and global change drivers”.
The purpose of this tutorial is to provide a detailed description of the modeling approaches used in the paper along with examples of the models in action on both simulated and real datasets.
Tools you’ll need
All models are Bayesian models run in R and JAGS. You will need these programs to run these models.
(We also recommend RStudio)
Other options are available for running Bayesian models, including Stan and NIMBLE. There is less flexibility but great usability for users of any experience level for running multi-species occupancy models in the package spOccupancy. Given the incorporation of uncertainty in the regression models we developed, it would be challenging to run them in programs with built-in regression model functions, but they can be built in any Bayesian modeling language/program (JAGS, Stan, BUGS, NIMBLE).
Model description
We employed a two-step modeling process (Figure 1).
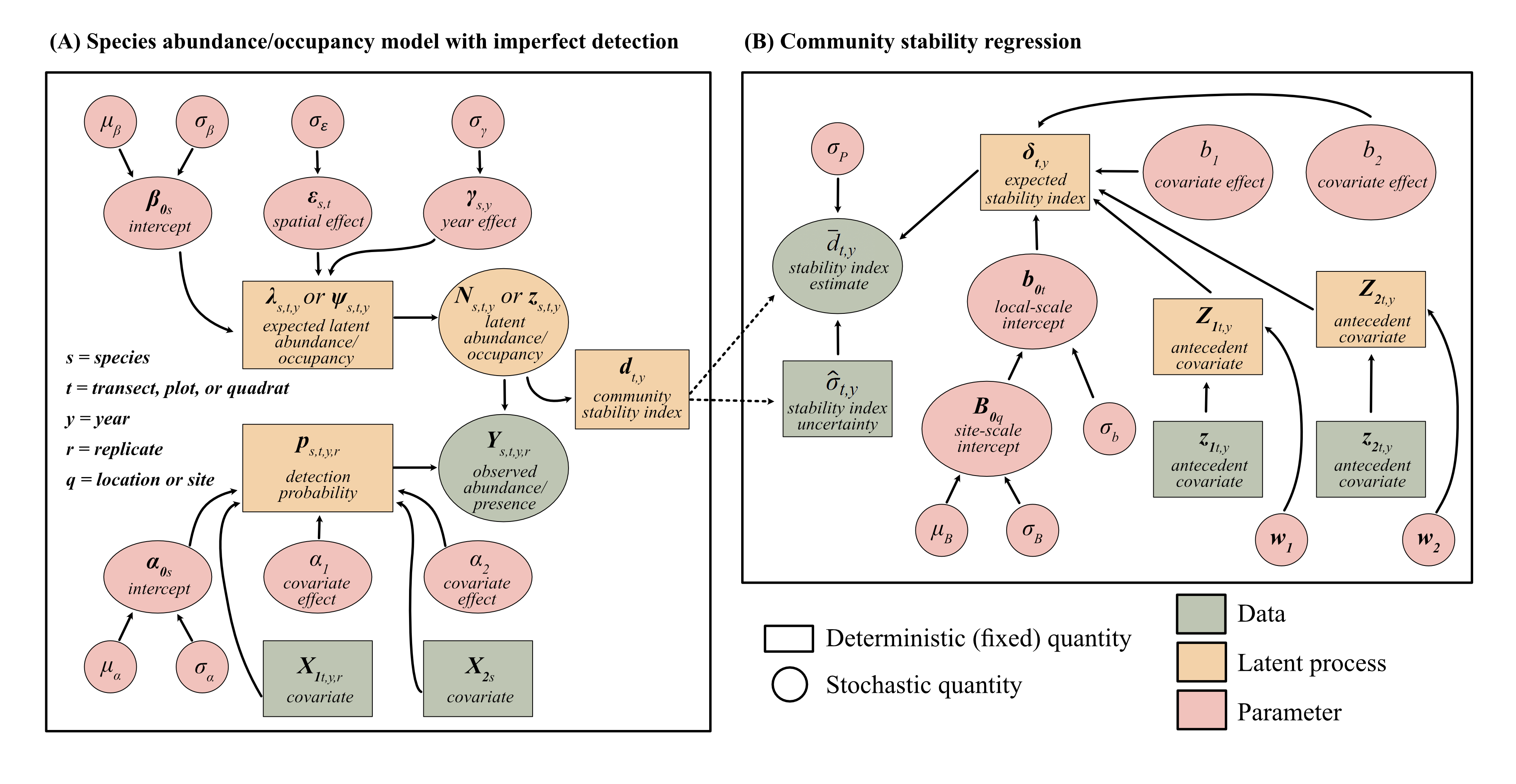
A) Multi-species model accounting for imperfect detection
This model links observed abundance or presence-absence of species in a community to detection probabilities to generate a latent “true” abundance or occupancy for each species in each site in each year. We used this model to then generate metrics of biodiversity. We built these models based on previous model developments for accounting for imperfect detection in community datasets (Dorazio et al. 2006).
B) Regression model with environmental covariates
This model incorporates mean and variance of biodiversity metrics from the model in A) into a regression examining the effects of environmental covariates on biodiversity. This model employs a stochastic antecedent modeling (SAM) framework (Ogle et al. 2015) to allow environmental covariates to have immediate and lagged responses.
Outline
In the following pages, we will walk you through the model formulation as well as model code demonstrating the modeling framework.
- Accounting for imperfect detection walks through accounting for imperfect detection in a multi-species model
- Computing indices of biodiversity illustrates how posterior samples for latent occupancy or abundance from the models for imperfect detection can be used to calculate multiple indices of biodiversity either inside Bayesian modeling software as derived quantities or through functions available in a variety of R packages
- Evaluating biodiversity in relation to environmental variables shows how to take these derived values of biodiversity and uncertainty around them and use them in a regression that allows covariates to have immediate and lagged effects on biodiversity metrics